Fig. 8
From: Picky with peakpicking: assessing chromatographic peak quality with simple metrics in metabolomics
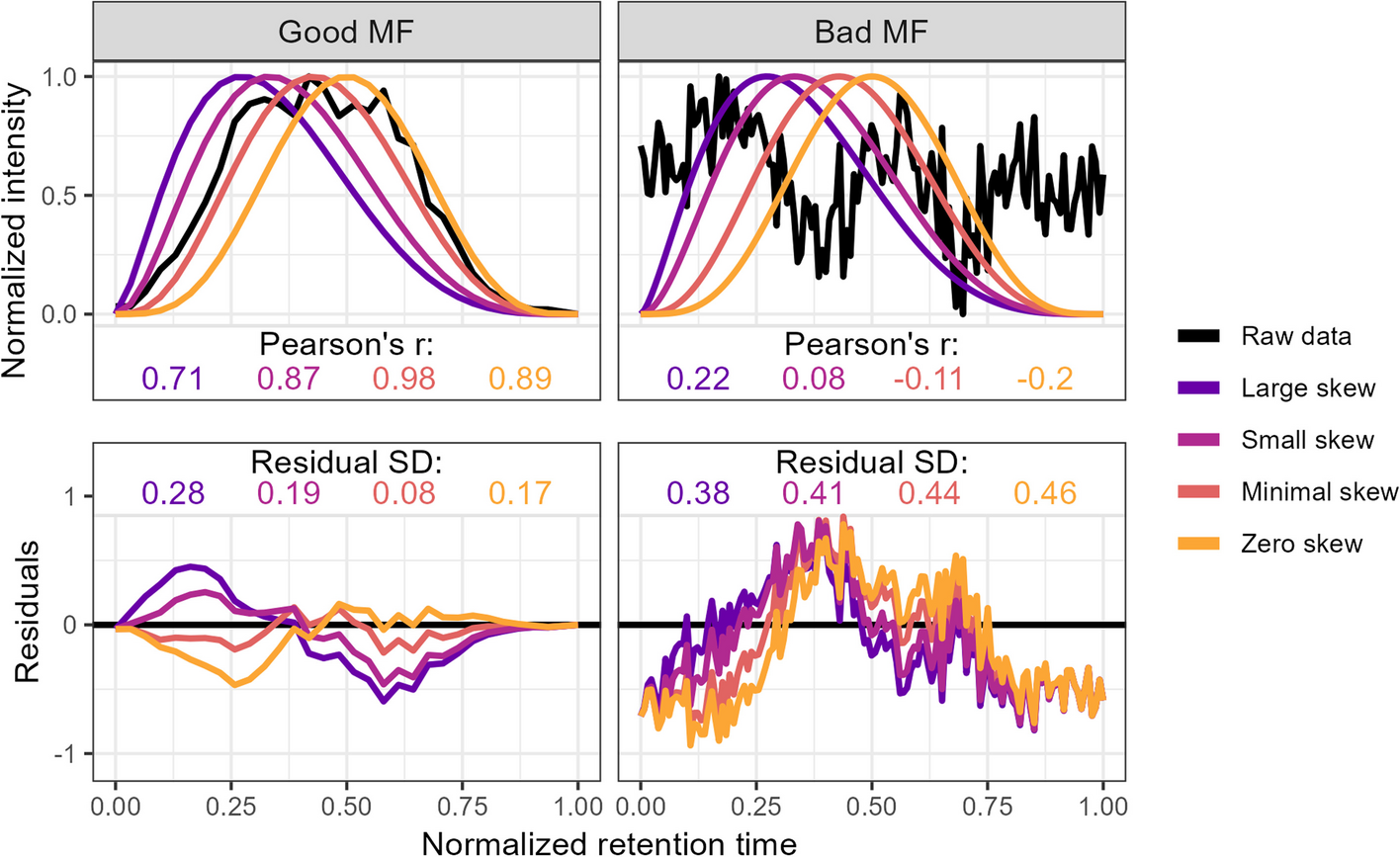
Method used to calculate the metrics for the two-parameter model from the raw data via comparison to an idealized pseudo-Gaussian peak for both manually identified “Good” and “Bad” peaks. Normalization was performed by linearly scaling the raw values into the 0–1 range by subtracting the minimum value and dividing by the maximum. Peak shape similarity was measured with Pearson’s correlation coefficient and the noise level is estimated as the standard deviation of the residuals after the raw data is subtracted from the idealized peak